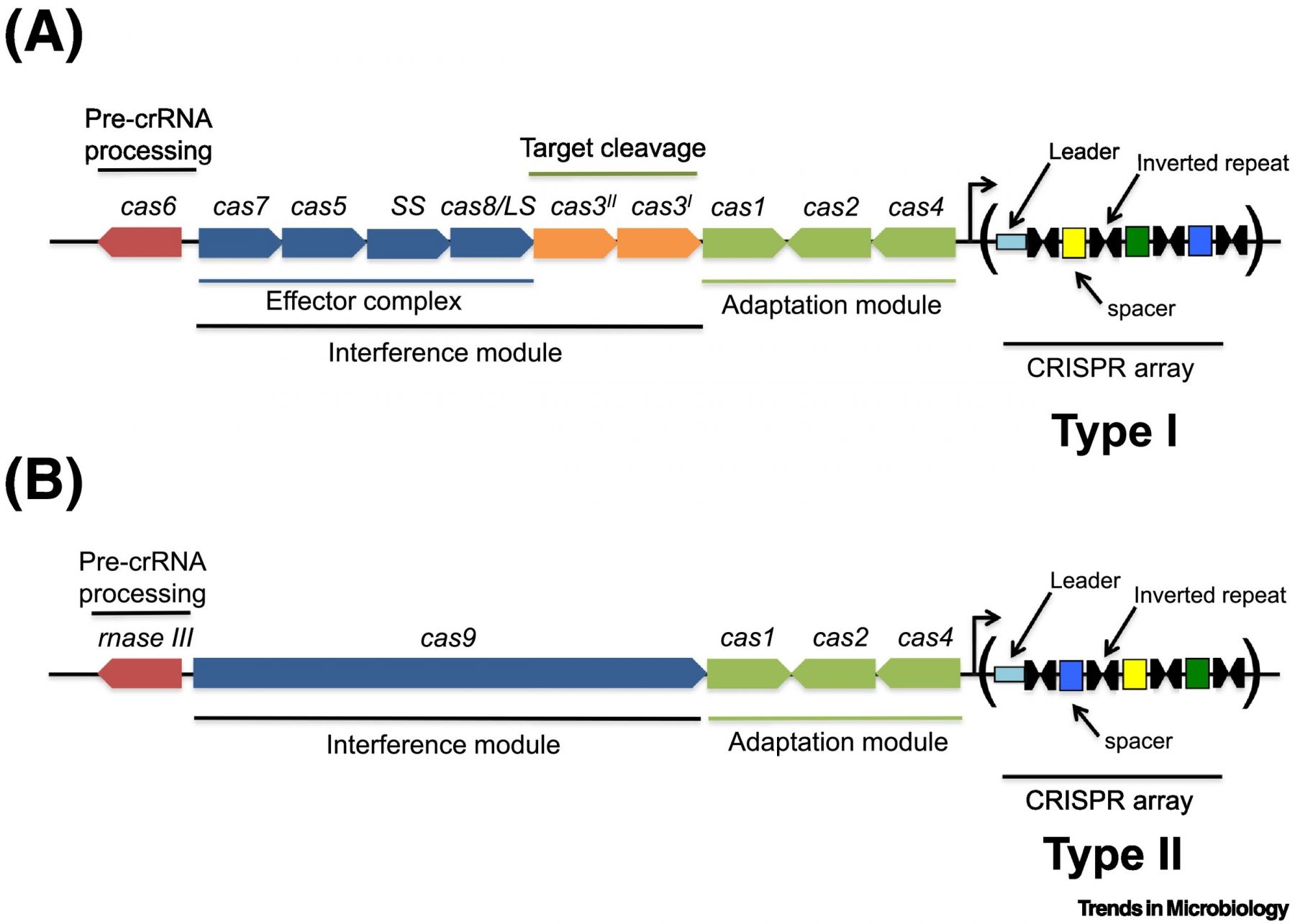
In this opinion article we highlight links between the H-NS nucleoid-associated protein, variable DNA topology, the regulation of CRISPR-cas locus expression, CRISPR-Cas activity, and the recruitment of novel genetic information by the CRISPR array. We propose that the requirement that the invading mobile genetic element be negatively supercoiled limits effective CRISPR action to a window in the bacterial growth cycle when DNA topology is optimal, and that this same window is used for the efficient integration of new spacer sequences at the CRISPR array. H-NS silences CRISPR promoters, and we propose that antagonists of H-NS, such as the LeuO transcription factor, provide a basis for a stochastic genetic switch that acts at random in each cell in the bacterial population. In addition, we wish to propose a mechanism by which mobile genetic elements can suppress CRISPR-cas transcription using H-NS homologues. Although the individual components of this network are known, we propose a new model in which they are integrated and linked to the physiological state of the bacterium. The model provides a basis for cell-to-cell variation in the expression and performance of CRISPR systems in bacterial populations.
Download “Article” CRISPR-Cas_Nucleoid_Associated_Proteins.pdf – Downloaded 401 times – 1 MB
Download a copy of the manuscript