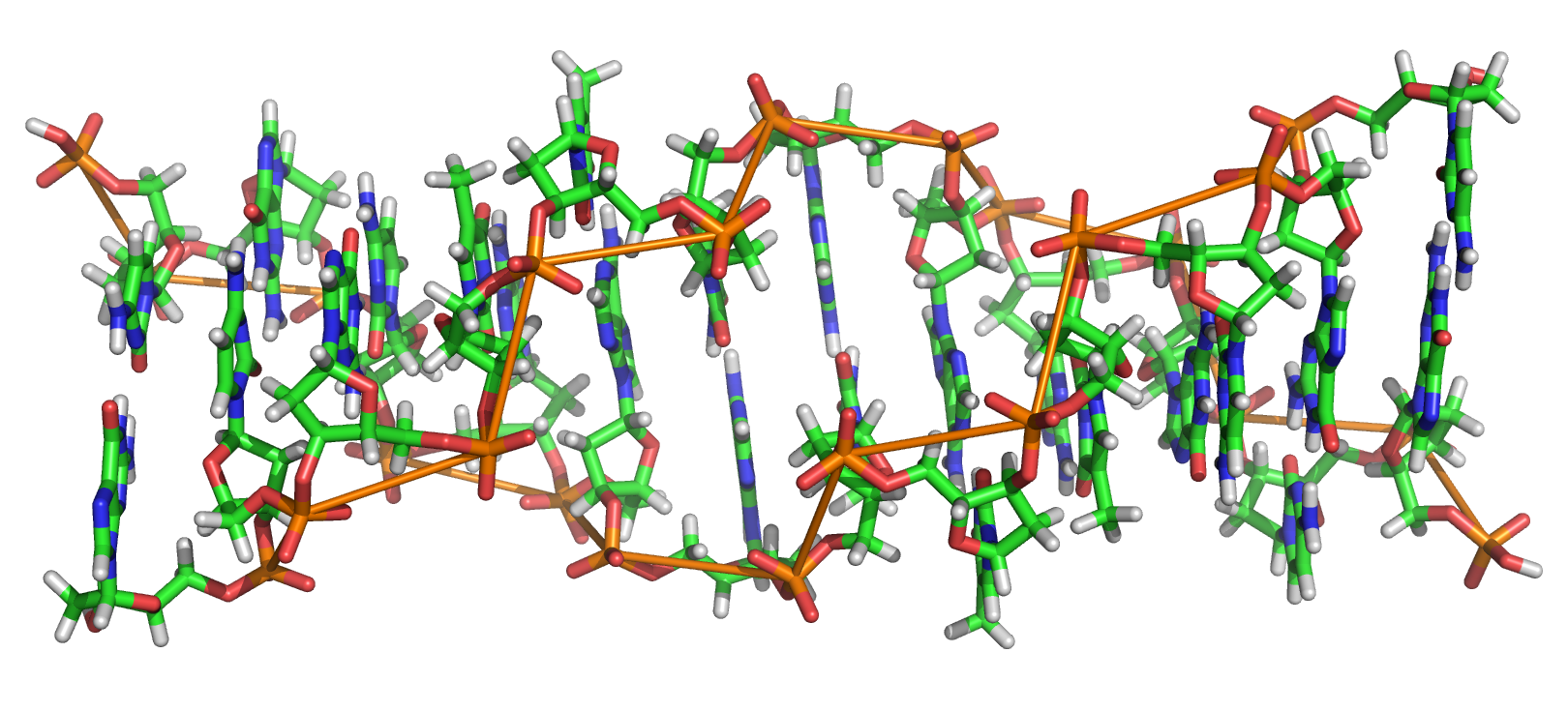
It is well known what sequences of bases in DNA are potentially in the left-handed form, i.e. prone to be in the Z-DNA conformation. In particular, d(GC)n > d(CA)n > d(CGGG)n > d(AT )n. These Z-DNA forming sequences (ZFS) have been found in the full DNA sequences of SARS CoV-2, rodent parvoviruses, salmonella, and some carcinogens. During a Short Term Scientific Mission (STSM, see the Introduction) we examined the stochastic profiles of such sequences in the FASTA format to determine the probability of these occurring. In the sequences studied we found CA more prevalent than GC, and also AT more common than CGGG. Novelly, we found such sequences in the Epstein-Barr virus (EBV), which to the best of our knowledge had not been thoroughly checked previously for ZFS, and we calculated the probabilities of those subsequences occurring. Note that in the EBV case, GC pairs were more prevalent than CA pairs. We also checked Dengue and HIV for potential ZFS and found many
potential sites in HV8. Finally, we present our current work, including implications for the 3-D conformation of the DNA molecule and applying the idea of microsatellites to the repeated sequences known to be in left-handed form, especially since these inform an analysis of the possible transition sites from the B-form of DNA to the Z-form, and vice versa. These transitions at CG pairs have been the most studied, but the flips seem to happen more at TG sites. AT pairs are also possible. As above, we find the probabilities of potential B-Z transitions at these locations using our methodology. Our results can help researchers hone in on the regions in genomes where Z-DNA formation is likely.
Download “Article” ZDNA.pdf – Downloaded 228 times – 2 MB
Download a copy of the manuscript