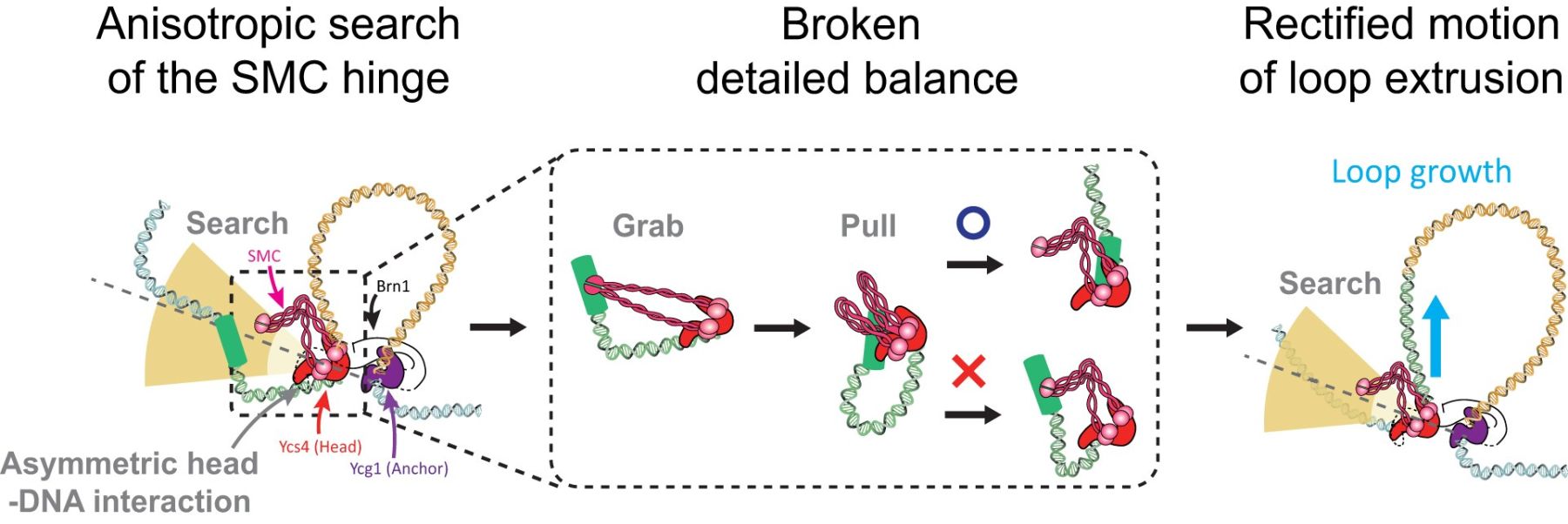
DNA loop formation by structural maintenance of chromosome (SMC) proteins, including cohesin, condensin, and the SMC5/6 complex, plays a pivotal role in genome organization. Despite its importance, the molecular mechanism underlying SMC-mediated loop formation, particularly how these complexes achieve persistent directionality (rectification) while minimizing backward steps during the formation of large loops, remains poorly understood. Here, we use atomic force microscopy (AFM) and computational simulation to uncover a key geometric feature of the yeast condensin SMC complex enabling rectified loop growth. Using AFM, we demonstrate that the hinge domain of yeast condensin exhibits a directional bias, extending orthogonally to the bound DNA and sampling an anisotropic region of space around the protein complex. By accounting for the geometric constraint on the hinge-mediated DNA-capture step, we computationally show that loop growth can spontaneously self-rectify. In contrast, an SMC model with broken detailed balance and isotropic search instead exhibited substantial loop shrinkage and random-walk-like behaviour. These findings reveal an overlooked, and potentially broadly conserved, anisotropic DNA capture mechanism through which SMC complexes form and stabilize DNA loops in vivo, in turn providing novel insights into the physical principles governing genome organization.
Download “Article” Spontaneously_directed_loop_extrusion.pdf – Downloaded 138 times – 3 MB
Download a copy of the manuscript