Leader: Davide Michieletto, University of Edinburgh, United Kingdom
Co-leader: Dorothy Buck, University of Bath, United Kingdom
Aleksandre Japaridze, Technical University of Delft, Netherlands
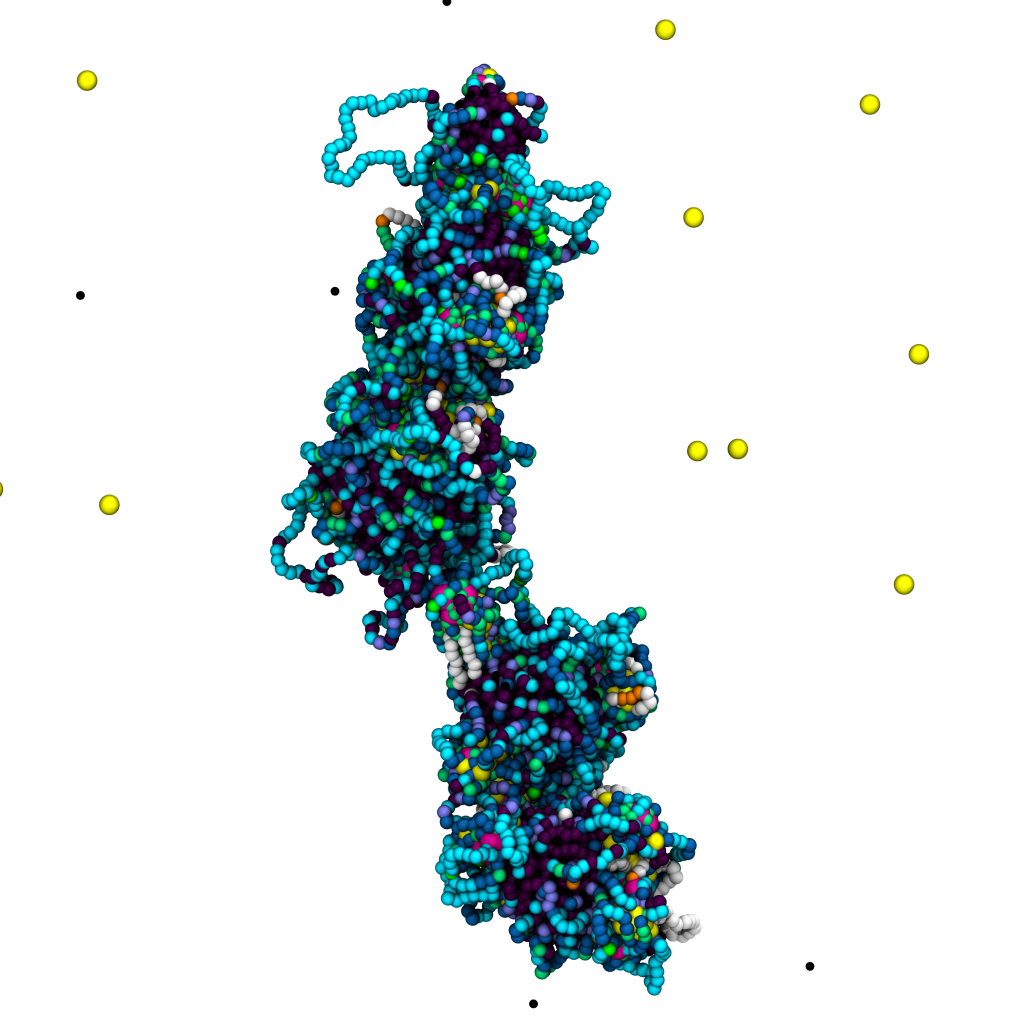
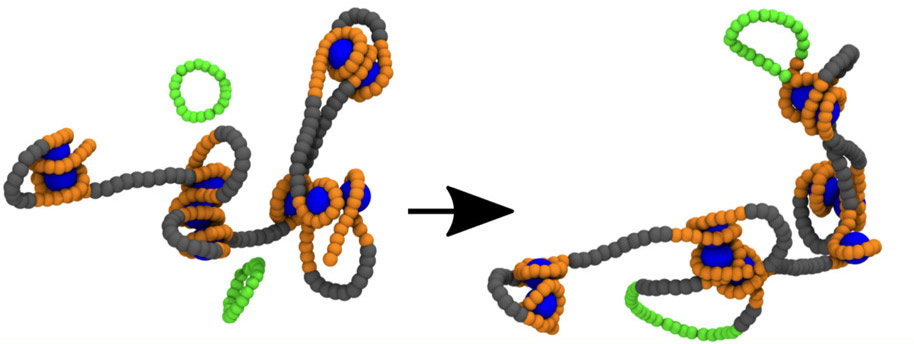
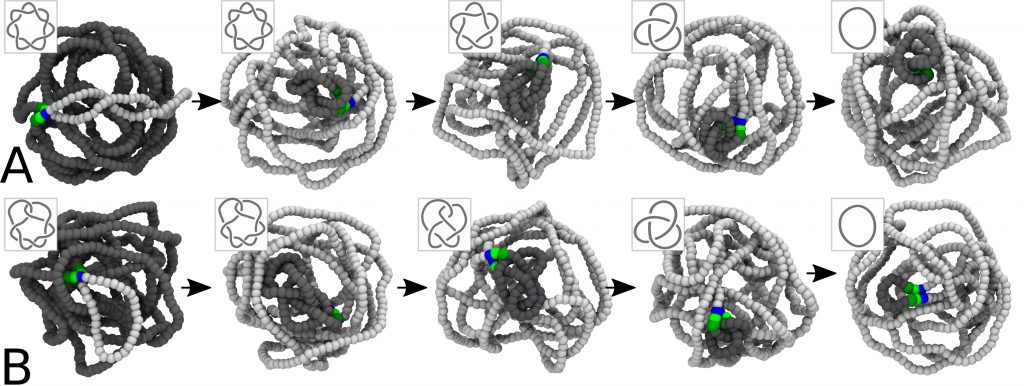
General overview Nature presents fascinating examples of genetic biomolecules and biomaterials ruled by topology: (1) The most “extreme” one is probably the DNA inside the cell nucleus, whose packing density makes it prone to attain an intricate, self-entangled structure. (2) The mithocondrial DNA of the organisms of the class kinetoplastida, called kinetoplast DNA (kDNA). This system is formed by thousands of DNA minicircles linked into a precise network, and is an important target of drugs aiming at killing the disease-carrying organism. (3) Viral DNA, whose level of confinement is so large that the biopolymer is often found heavily knotted. In this context, novel and powerful experimental methods have just started to unveil the strong link between topological arrangement and biological function. Yet, the interpretation of these experimental data appears often problematic, hence posing the key conceptual challenge of the next few years: it requires, in fact, expertise in generic topological problems in polymers, computational and theoretical physics, as well as molecular and cell biology/biochemistry.
General objectives of WG4. DNA represents one of the core systems whose topological properties will be studied in the course of the proposed EUTOPIA Action. Specifically, two forms of DNA-based materials will be at the centre of the Network’s interest, namely kinetoplast DNA and chromatin. The evolution, formation and replication of kinetoplast DNA is a largely unsolved mystery, and presents many opportunities for technological applications. Taking inspiration from the topological and elastic properties of the kDNA, researchers in EUTOPIA aim at studying models of networks of randomly linked rings with non standard mechanical properties. An important issue that will also be explored is the effect that confinement can have on the topological properties of the formed networks and use the results obtained on this controlled system to infer about properties of nuclear DNA. Chromatin, which is better known than kDNA, shows the tendency to be organized hierarchically, with small domains packed close-by to form domains of larger size and so forth. This particular organization has a profound impact on processes like gene expression and regulation, yet how topology directly influences its formation remains a mystery which EUTOPIA aims to address in great detail. The Action will push the boundaries of what is currently known about these systems by:
- Performing simulations of ring polymer networks varying concentration, stiffness, and type of topological link that can occur among rings.
- Identifying the pertinent variables and analyse the simulations results to get general dependencies between physical and topological parameters.
- Pushing on the development of more refined computational models of DNA, chromatin fibres, and chromosomes, capable of accounting for the intrinsic multi-scale nature of the problem.
- Developing numerical and analytical technologies to account for topological properties in simulation models.
- Employing these methodologies to explore and predict the influence of ‘active’ mechanisms on DNA and chromatin topologies within the crowded environment of the nucleus.
Specific tasks of WG4. Due to their size and the strong coupling between small- and large-scales, DNA filaments and chromatin remain untreatable for all-atom simulations even by modern super-computers. The role of this WG concerns the development of theoretical and computational methodologies for investigating the spatio-temporal behaviour of DNA and chromatin in “extreme”, topology-sensitive environments such as the nucleus of the eukaryote cell or the bacterial nucleoid. In order to realize this goal, the following inter-connected tasks have been identified:
- Development of systematic coarse-grain strategies for DNA and chromatin, with particular emphasis on computational models capable of matching between different resolutions in space and time (multi-scale models).
- Exploration of the relationship between topology and functions of DNA and chromatin in various biological contexts.
- Development of on-line tools for the analysis of three-dimensional DNA and chromatin organization based on the computational tools developed in Task 1. These tools should become “the” standard for the whole community. Relevant for WG1.
- Modelling of sequencing and other experimental techniques to assess the impact of topological entanglement on their reliability, and to support the design of efficient sequencing devices.
- Explore the interplay of active and passive mechanisms controlling the entanglement of chromatin, and nucleic acids in general, within the crowded nuclear environment.
Deliverables: STSMs within the WG and to other WGs. At least one article with 3 international members per each task. Instalment of collaborations with the industry and with biological labs.