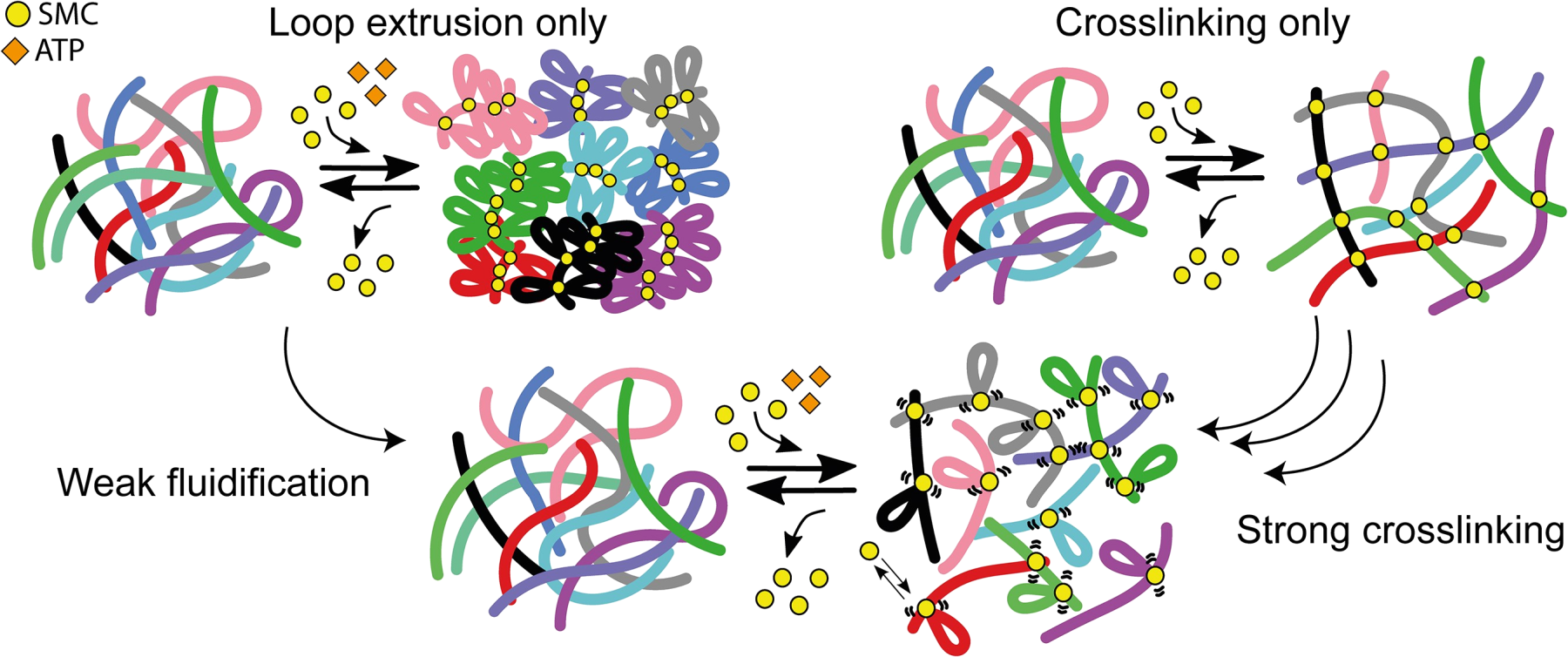
Structural-Maintenance-of-Chromosome (SMC) complexes, such as condensins, organise the folding of chromosomes. However, their role in modulating the entanglement of DNA and chromatin is not fully understood. To address this question, we perform single-molecule and bulk characterisation of yeast condensin in entangled DNA. First, we discover that yeast condensin can proficiently bind double-stranded DNA through its hinge domain, in addition to its heads. Through bulk microrheology assays, we then discover that physiological concentrations of yeast condensin increase both the viscosity and elasticity of dense solutions of λ-DNA, suggesting that condensin acts as a crosslinker in entangled DNA, stabilising entanglements rather than resolving them and contrasting the popular theoretical picture where SMCs purely drive the formation of segregated, bottle-brush-like chromosome structures. We further discover that the presence of ATP fluidifies the solution–likely by activating loop extrusion–but does not recover the viscosity measured in the absence of protein. Finally, we show that the observed rheology can be understood by modelling SMCs as transient crosslinkers in bottle-brush-like entangled polymers. Our findings help us to understand how SMCs affect the dynamics and entanglement of genomes.
Download “Article” Yeast_condensin_acts.pdf – Downloaded 20 times – 4 MB
Download a copy of the manuscript