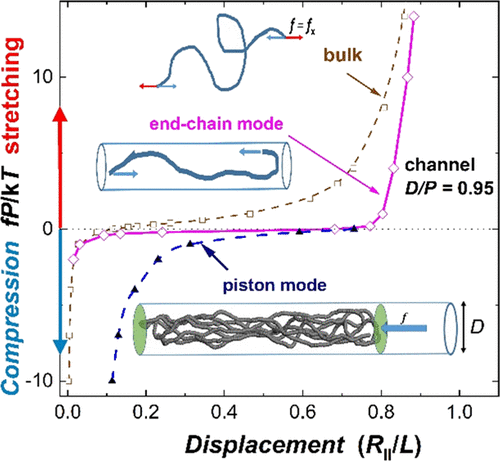
We study the compression and extension response of single dsDNA (double-stranded DNA) molecules confined in cylindrical channels by means of Monte Carlo simulations. The elastic response of micrometer-sized DNA to the external force acting through the chain ends or through the piston is markedly affected by the size of the channel. The interpretation of the force (f)–displacement (R) functions under quasi-one-dimensional confinement is facilitated by resolving the overall change of displacement ΔR into the confinement contribution ΔRD and the force contribution ΔRf. The external stretching of confined DNA results in a characteristic pattern of f–R functions involving their shift to the larger extensions due to the channel-induced pre-stretching ΔRD. A smooth end-chain compression into loop-like conformations observed in moderately confined DNA can be accounted for by the relationship valid for a Gaussian chain in bulk. In narrow channels, the considerably pre-stretched DNA molecules abruptly buckle on compression by the backfolding into hairpins. On the contrary, the piston compression of DNA is characterized by a gradual reduction of the chain span S and by smooth f–S functions in the whole spatial range from the 3d near to 1d limits. The observed discrepancy between the shape of the f–R and f–S functions from two compression methods can be important for designing nanopiston experiments of compaction and knotting of single DNA in nanochannels.
Download “Article” Compression_Stretching_Single_DNA_Molecules.pdf – Downloaded 353 times – 2 MB
Download a copy of the manuscript