Status
Completed
Period
13 January – 13 March 2020
Applicant
Dr Marco Di Stefano
Home Institution
Postdoctoral researcher at CNAG-CRG, Centre for Genomic Regulation (CRG), Barcelona Institute of Science and Technology (BIST), Carrer Baldiri i Reixac 4, 08028 Barcelona, Spain.
Host Contact
Dr Daniel Jost
Host Institution
Ecole Normale Superieure de Lyon, Laboratory of Biology and Modeling of the Cell, Parvis Rene Descartes 15, 69342, Lyon, France
Aim of the mission
In the plant Arabidopsis thaliana, several experimental studies have characterised the genome spatial organization. Imaging revealed that (i) chromosomes are organized in territories (Fransz et al. 2002) , (ii) the nucleolus generally localizes at the nuclear centre (Pontvianne et al. 2016) , (iii) telomeres cluster at the nucleolar periphery and (iv) centromeres are positioned at the nuclear periphery (Fransz et al. 2002; Fang and Spector 2005) . Complementarily, Hi-C experiments revealed the presence of specific interaction patterns: (i) strong inter- and intra-chromosome interactions between heterochromatic regions (Grob, Schmid, and Grossniklaus 2014; Feng et al. 2014) , and (ii) positive stripes of frequent interactions with neighboring chromatin regions enriched with H3K27me3, a histone modification associated with Polycomb repressive complexes (Wang et al. 2015) .
In this STSM, we aim to perform theoretical studies to complement the experimental findings and try to unravel the mechanisms that are driving the genome organization. We will build coarse-grained genome-wide models of the A. thaliana nucleus and test to which extent the structure of the genome organization may be related to the epigenetic states of the chromatin. Specifically, we will explore the interplay of two mechanisms: topological restraints, controlling the entanglement of chromatin within the nuclear environment, and the attraction or repulsion between loci depending on their epigenetic state. We will, then, validate quantitatively our predictions against published experimental Hi-C and imaging data.
Summary of the results
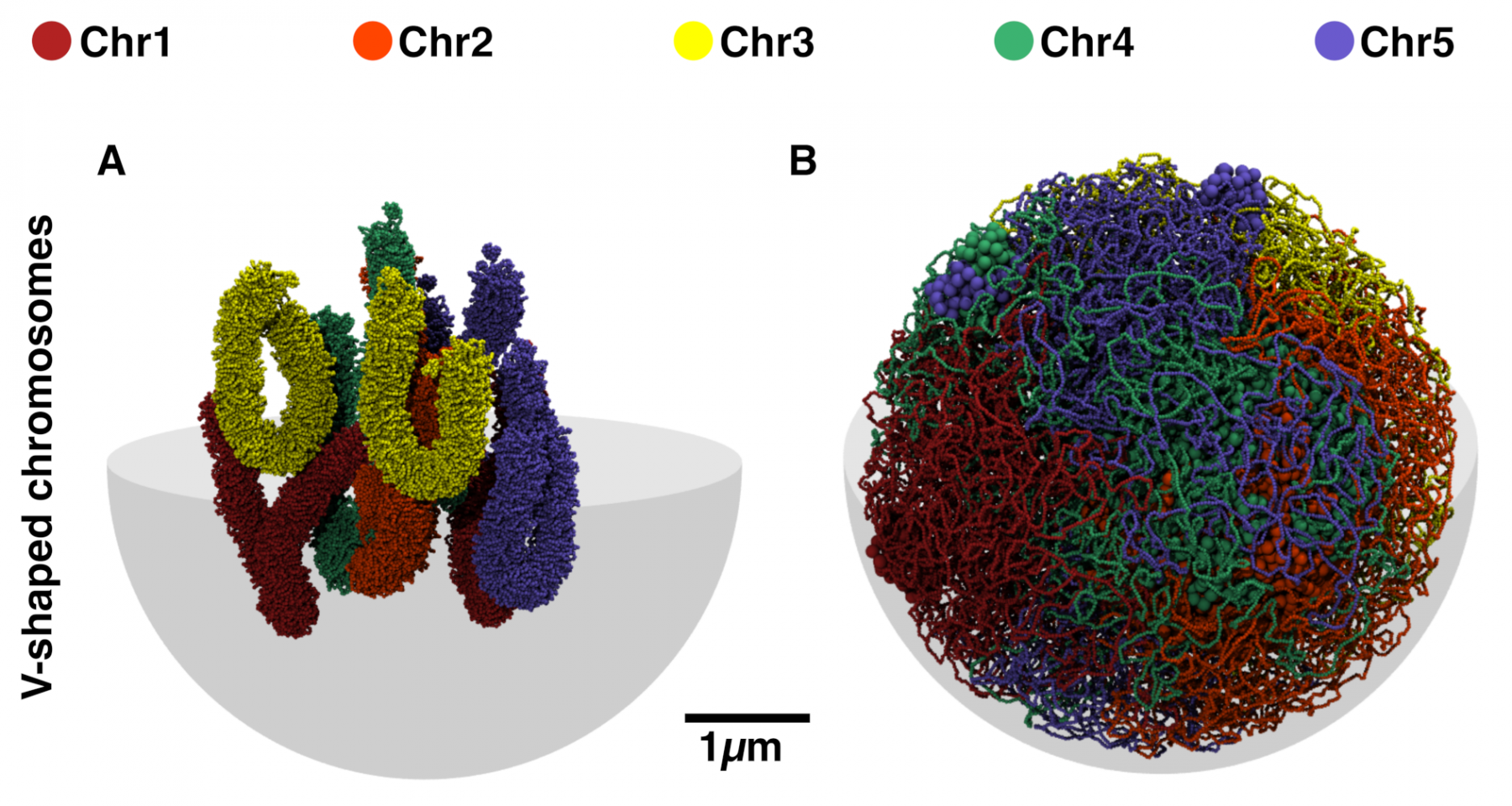
We found that the initial chromosome conformation is of paramount importance to predict the experimental data. Due to topological constraints, organizing chromosomes as V-shaped objects give higher correlations with the Hi-C experimental data than linear rod-like arrangements. Additionally, the self-attraction of the nucleolar organizing regions (NORs) have a crucial role in the formation and the maintenance of the nucleolus at the nuclear centre. The repulsion between constitutive heterochromatin (CH) and the other epigenomic states allow explaining the segregation of the CH-rich chromocenters, their effective self-attraction, and their preferential localization at the nuclear periphery.
Dissemination
During the STSM, we almost completed the writing of a manuscript describing the models and our main results. The manuscript is now available here. The work and the discussions with all the members of the host lab opened new exciting venues. Hopefully, they will lead to continuing the collaboration on chromatin modelling with the visitor. During the STSM, the visitor also provided a training session for the host laboratory on TADbit, a Python library for the bioinformatic analysis of Hi-C datasets which he contributed to developing.
References
Fang, Yuda, and David L. Spector. 2005. Molecular Biology of the Cell . https://doi.org/ 10.1091/mbc.e05-08-0706 .
Feng, Suhua, Shawn J. Cokus, Veit Schubert, Jixian Zhai, Matteo Pellegrini, and Steven E. Jacobsen. 2014. Molecular Cell 55 (5): 694–707.
Fransz, Paul, J. Hans De Jong, Martin Lysak, Monica Ruffini Castiglione, and IngoSchubert. 2002. Proceedings of the National Academy of Sciences of the United States of America 99 (22): 14584–89.
Grob, Stefan, Marc W. Schmid, and Ueli Grossniklaus. 2014. Molecular Cell . https://doi.org/ 10.1016/j.molcel.2014.07.009 .
Pontvianne, Frédéric, Marie-Christine Carpentier, Nathalie Durut, Veronika Pavlištová, Karin Jaške, Šárka Schořová, Hugues Parrinello, et al. 2016. Cell Reports 16 (6): 1574–87.
Wang, Congmao, Chang Liu, Damian Roqueiro, Dominik Grimm, Rebecca Schwab, Claude Becker, Christa Lanz, and Detlef Weigel. 2015. Genome Research . https://doi.org/ 10.1101/gr.170332.113 .