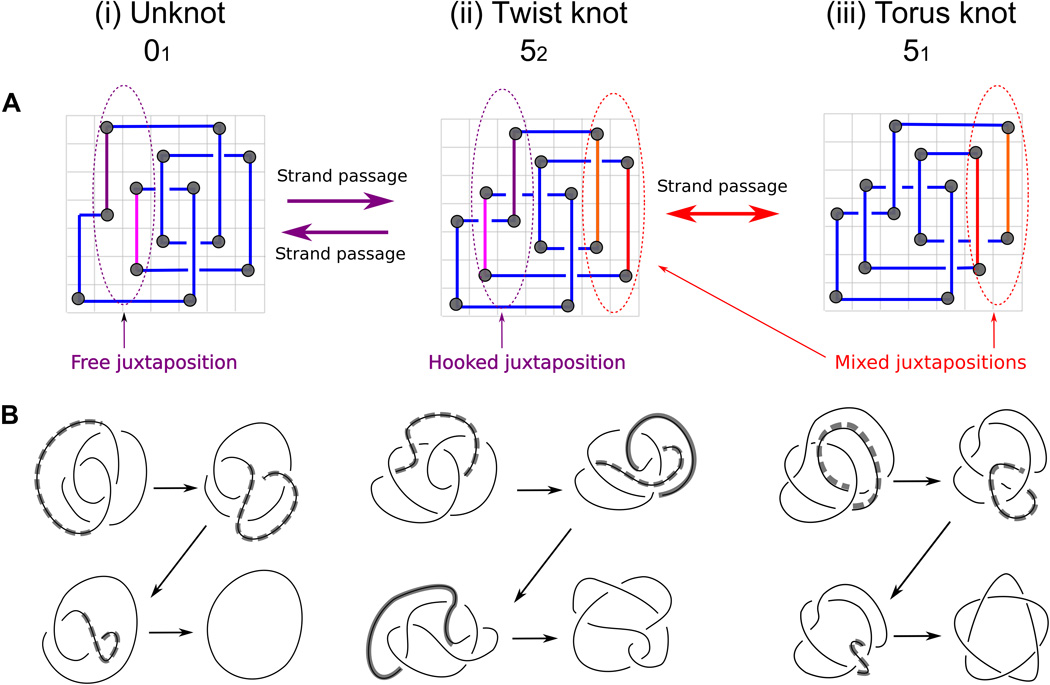
Grid diagrams with their relatively simple mathematical formalism provide a convenient way to generate and model projections of various knots. It has been an open question whether these 2D diagrams can be used to model a complex 3D process such as the topoisomerase-mediated preferential unknotting of DNA molecules. We model here topoisomerase-mediated passages of double-stranded DNA segments through each other using the formalism of grid diagrams. We show that this grid diagram–based modeling approach captures the essence of the preferential unknotting mechanism, based on topoisomerase selectivity of hooked DNA juxtapositions as the sites of intersegmental passages. We show that the grid diagram–based approach provides an important, new, and computationally convenient framework for investigating entanglement in biopolymers.
Download “Article” grid_diagrams_DNA_topology.pdf – Downloaded 355 times – 1 MB
Download a copy of the manuscript