Status
Completed
Period
13-19 February 2023
Applicant
Giovanni Mattiotti
Home Institution
Department of Physics, University of Trento
Host Contact
Prof. Samuela Pasquali
Host Institution
Unité de Biologie Fonctionnelle & Adaptative, Université de Paris, CNRS UMR 8251
Aim of the mission
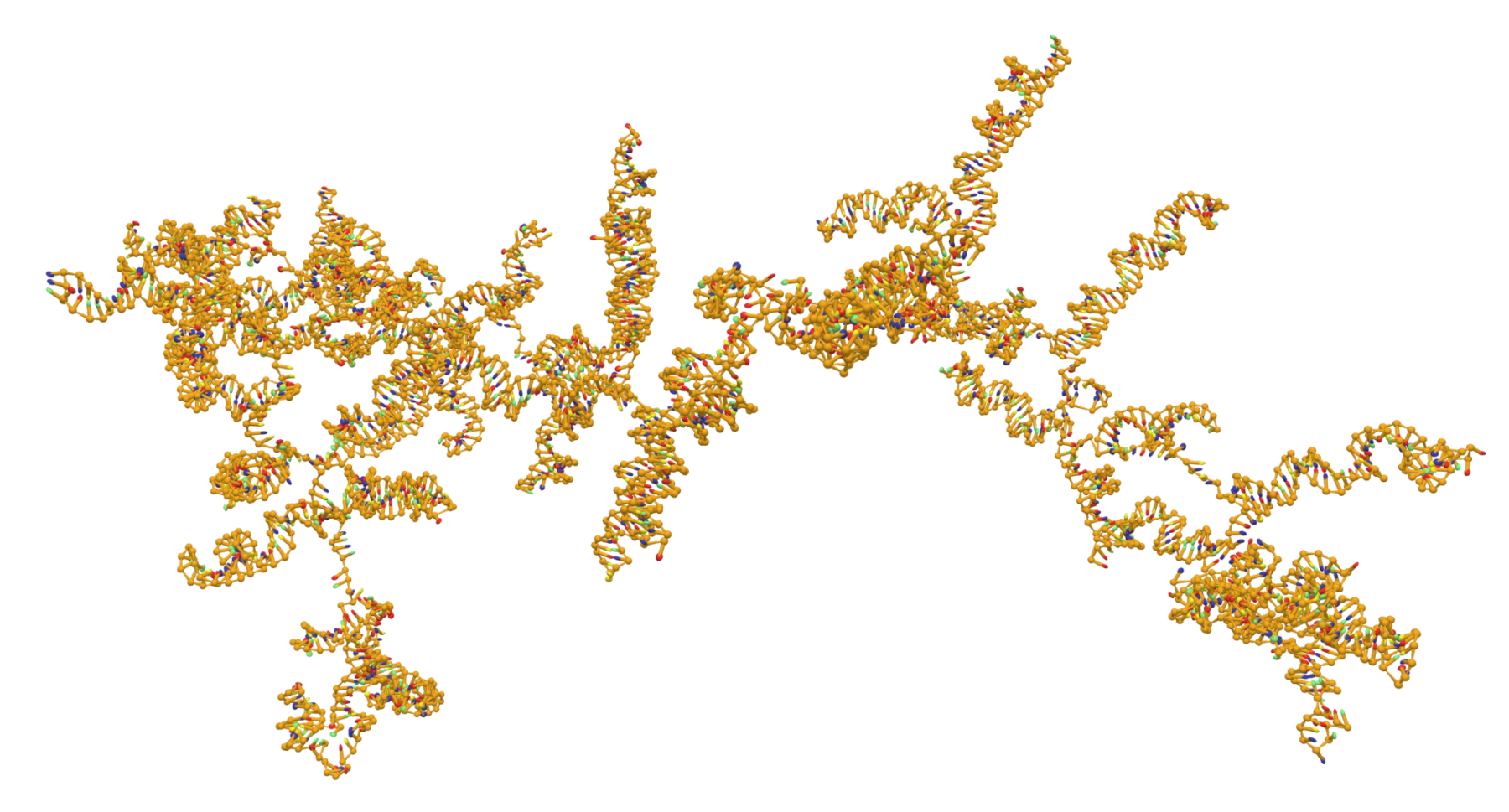
The process of self-assembly (SA) of biomolecular structures, such as viral particles, is a fascinating topic whose general understanding impacts several scientific fields, from evolutionary biology to drug delivery. However, its intrinsic complexity poses a challenge for the high-resolution reconstruction of the assembly pathway, both to experimental and computational scientists. While models and simulations have provided extremely insightful spatial and temporal details, they are limited by the currently available computational power. On the other hand, in vitro experiments are able to reproduce the process in a more realistic way, but they cannot return the chemical detail that is essential to clarify the mechanisms involved. Leveraging the theoretical efficiency provided by a recently developed coarse-grained (CG) model of RNA (oxRNA [1]), we have decided to assess the folding pathways of a ssRNA viral fragment in the context of the SA process by means of molecular dynamics (MD) simulations. By construction, the oxRNA model has been developed to consistently predict the emergence of secondary and tertiary structural motifs, in spite of allowing for the sole Watson-Crick coupling of base pairs. In addition, the level of resolution of the model is suitable to perform an extensive exploration of the configurational space of biologically-relevant systems, expanding on the typical timescales of classic MD assessments by more than 3 orders of magnitude with respect to all-atom (AA) simulations. Therefore, simulations performed with the oxRNA force field might provide insightful details on the earliest stages of SA processes, which have been shown in vitro to occur on timescales about the order of seconds [2].
The goal of our project is twofold:
1. To give an extensive description of the conformational space of the RNA2 fragment found in the CCMV viral capsid both in the presence and in the absence of an external field that mimics the effect of the capsid itself;
2. to produce structures with realistic topologies and shapes to be back-mapped into AA representations of the system, which would be subsequently employed in high-resolution simulations of the whole virion, i.e. the CCMV proteic capsid containing the RNA2 fragment.
An insightful feature of these simulations would be the spontaneous emergence of secondary and tertiary structural motifs in the viral RNA, tracked via the evolution of the hydrogen-bond network between nucleotides. Another critical aspect of this investigation would be the detection of knots: indeed, we expect to observe substantial topological differences by enforcing physical constraints, such as by imposing an external field confining the nucleic acid chain in a restricted volume [3].
Even though a significant part of the work has already been carried out, a substantial amount of simulations, as well as a thorough analysis of the raw data, remains to be performed. For these reasons, I would strongly benefit from a direct interaction with professor Samuela Pasquali (host of this STSM). She is a leading expert in the computational study of the folding pathways of single- stranded RNA filaments, specifically concerning the employment and development of alternative RNA force fields [4] and of advanced, ad hoc analysis techniques. By proposing a cutting-edge scenario that effectively pushes the boundaries of both the modeling and the analysis of complex RNA systems, this STSM is expected to lead the way of a prolific scientific collaboration, aiming to (at least) one major scientific publication.
[1] Petr Sulc et al. “A nucleotide-level coarse-grained model of RNA”. In: The Journal of Chemical Physics 140.23 (2014), p. 235102. doi: 10.1063/1.4881424. eprint: https://doi.org/10. 1063/1.4881424. url: https://doi.org/10.1063/1.4881424.
[2] Rees F. Garmann, Aaron M. Goldfain, and Vinothan N. Manoharan. “Measurements of the self-assembly kinetics of individual viral capsids around their RNA genome”. In: Proceedings of the National Academy of Sciences 116.45 (2019), pp. 22485–22490. doi: 10.1073/pnas. 1909223116. eprint: https://www.pnas.org/doi/pdf/10.1073/pnas.1909223116. url: https://www.pnas.org/doi/abs/10.1073/pnas.1909223116.
[3] Cristian Micheletti, Davide Marenduzzo, and Enzo Orlandini. “Polymers with spatial or topo- logical constraints: Theoretical and computational results”. In: Physics Reports 504.1 (2011), pp. 1–73. issn: 0370-1573. doi: https://doi.org/10.1016/j.physrep.2011.03.003. url: https://www.sciencedirect.com/science/article/pii/S0370157311000640.
[4] Samuela Pasquali and Philippe Derreumaux. “HiRE-RNA: A High Resolution Coarse-Grained Energy Model for RNA”. In: The Journal of Physical Chemistry B 114.37 (2010). PMID: 20795690, pp. 11957–11966. doi: 10.1021/jp102497y.
Summary of the Results
The goal of this short-term scientific mission was to build a pipeline of analysis to perform onto coarse-grained and all-atom structures of large RNA systems, with a focus on highlighting topological features such as pseudo-knots. Additionally, we aimed to gain a deeper understanding of simulating this type of system under more realistic conditions. I am happy to report that the STSM successfully achieved its planned goals and expected outcomes. Throughout the mission, we worked diligently to develop and refine the analysis pipeline. We incorporated a variety of techniques and tools, and were able to accurately identify topological features in the RNA systems we studied. In addition to the analysis work, we also devoted significant time to simulating these systems under more realistic conditions. This involved a great deal of experimentation, and we were ultimately able to develop an effective simulation approach that allowed us to study these systems with a high degree of accuracy and precision.
Dissemination
Moving forward, we have identified a number of potential future collaborations that would build on the work we did during the STSM. One opportunity is the post-doctoral position that Professor Pasquali will begin in November 2023. Thanks to our collaboration during the STSM, we were able to build a strong professional relationship and get to know each other well. This experience has motivated me to apply for the position, and I look forward to the possibility of working together again in the future. Overall, I believe that the STSM was an excellent opportunity to push the boundaries of our understanding of large RNA systems. By building a robust analysis pipeline and developing new simulation approaches, we were able to gain new insights into the structure and function of these complex systems. In particular, I want to underline the fact that there is a strong interest in studying large ssRNA structures, among the participants of the Action EUTOPIA. On the other hand, my contribution is orthogonal to the theoretical and experimental approaches followed by the other members: thanks to this mission I was able to advance in this almost unexplored direction of simulating large ssRNA systems with molecular dynamics, and the results I will get from my work will certainly be of interest for the entire community of the Action.