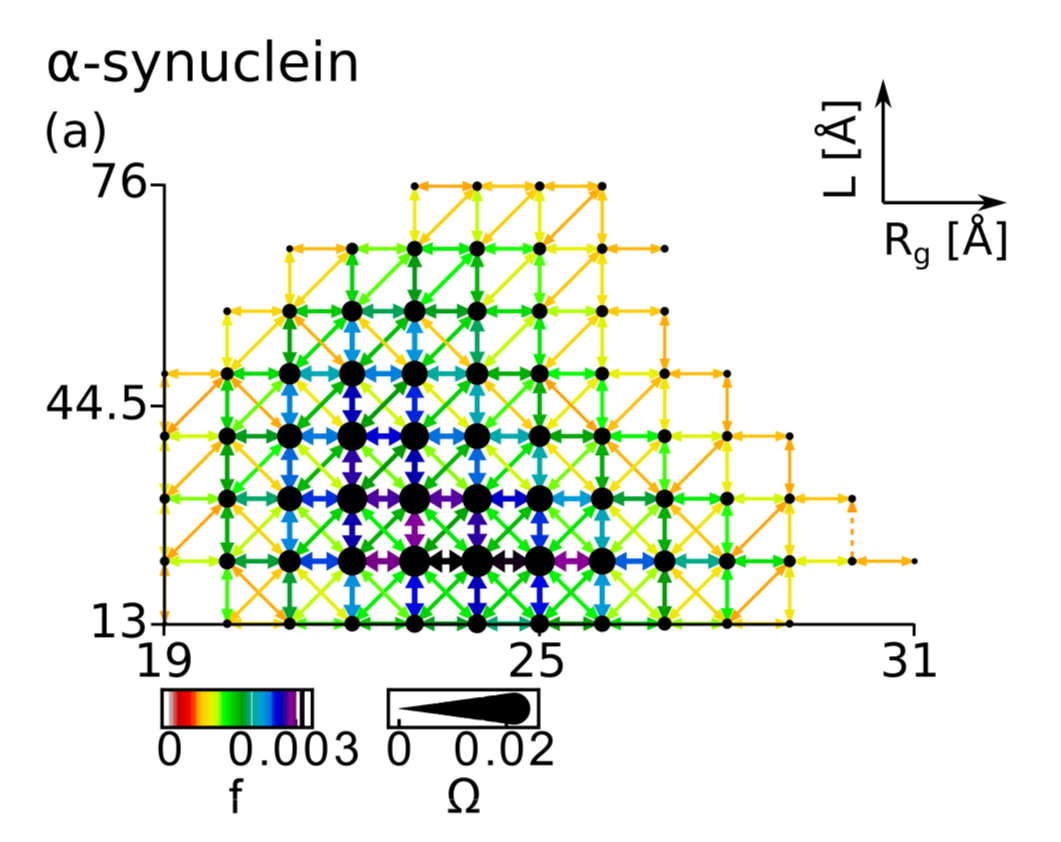
The equilibrium dynamics of the intrinsically disordered proteins is thought to consist of transitions between many basins in the free energy landscape whereas structured proteins stay in the vicinity of one native basin. We demonstrate this picture explicitly by studying networks defined on the discretized plane: conformational end-to-end distances vs radii of gyration. The bin sizes are defined by time scales that span orders of magnitude. The networks, derived from all-atom and coarse-grained molecular dynamics simulations, are nearly scale invariant. The bin representation also provides insights into the folding process of the structured proteins and identifies regions of hindrance to folding.
Download a copy of the manuscript