Retroviral infection is a process through which a virus injects its genetic material (RNA) into a host cell. The RNA is then retro-transcribed into DNA, which is then “integrated” into the host genome. The most famous example of retrovirus is HIV.
The way in which HIV and other retroviruses infect the host is still an open problem. Intriguingly, the DNA of retroviruses is frequently found in specific regions of genomes called “hot-spots” which display highly non-random distributions. The process underlying this non-random integration site selection has been subject of decades of intense research and debate. Most of this phenomenology is reported as empirical observations and attributed to the action of proteins, such as chaperones.
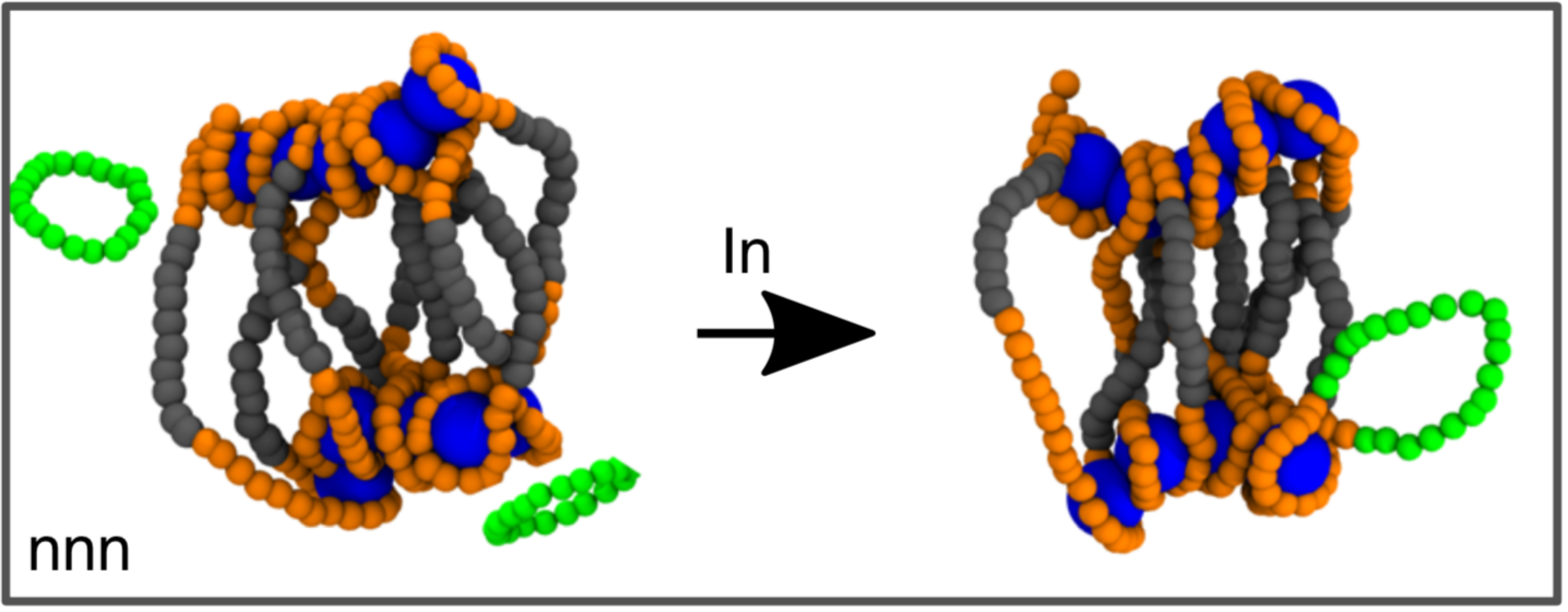
At the same time, the integration of HIV and other viral DNA into the host is a topological problem, in that it requires a reconnection operation to be performed in order to permanently insert the viral genetic material (topologically equivalent to a circle) into host chromosomes (topologically equivalent to a line).
In this paper, we propose that the bias in integration site selection may be partially due to previously unappreciated physical mechanisms. To test our hypothesis, we developed a biophysical model for retroviral integration as stochastic and quasi-equilibrium topological reconnections between polymers that are transiently proximal in 3D.
We discovered that physical effects, such as DNA accessibility and elasticity, play important and universal roles in this process and our simulations can generate predictions in quantitative agreement with experiments done in the last three decades on HIV integration in the human genome.
Most importantly, this work will pave the way for more experiments and modelling work thanks to the marriage between two previously disconnected fields, that of polymer topology and HIV infection.
Download a copy of the manuscript (preprint version)