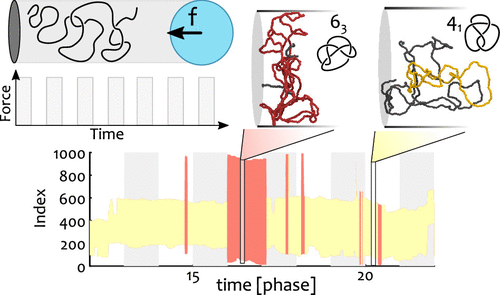
How do knots form and transform, even in our ordinary shoelaces and headphones, is puzzling and not fully understood. A similar problem is found in biology, where DNA is strongly packaged inside cells and so supposedly heavily knotted (although recent evidence suggest otherwise). To better understand the formation of knots, in DNA but also generic polymers, researchers have employed cutting-edge technologies where DNA is confined within thin cylinders and compressed by “nano dozers” — basically a piston that makes DNA crumple onto itself — and study the emergence and dissipation of knots and other entanglements. In spite of the beauty of these experiments, much of the information is obscured by the fact that optical microscopes cannot resolve DNA strands that are closer than 200 nm; in turn, this implies that a precise characterisation of the type of entanglement formed in these “nano dozer” experiments is not possible. To overcome this limitation, here we employ computer simulations and advanced topological profiling algorithms to mimic the nano-dozer experimental set up and fully characterise knots and entanglements in polymers that are pushed out of equilibrium. Our key discovery is that knots are surprisingly uncoupled from the external driving force and “have a life of their own”. On the contrary, so-called geometrical entanglement — or the self-crossing of the polymer onto itself — not only closely follows the modulation of the driving force but can also be finely tuned by adjusting the magnitude and frequency of the compressions.
Download a copy of the manuscript (preprint version)