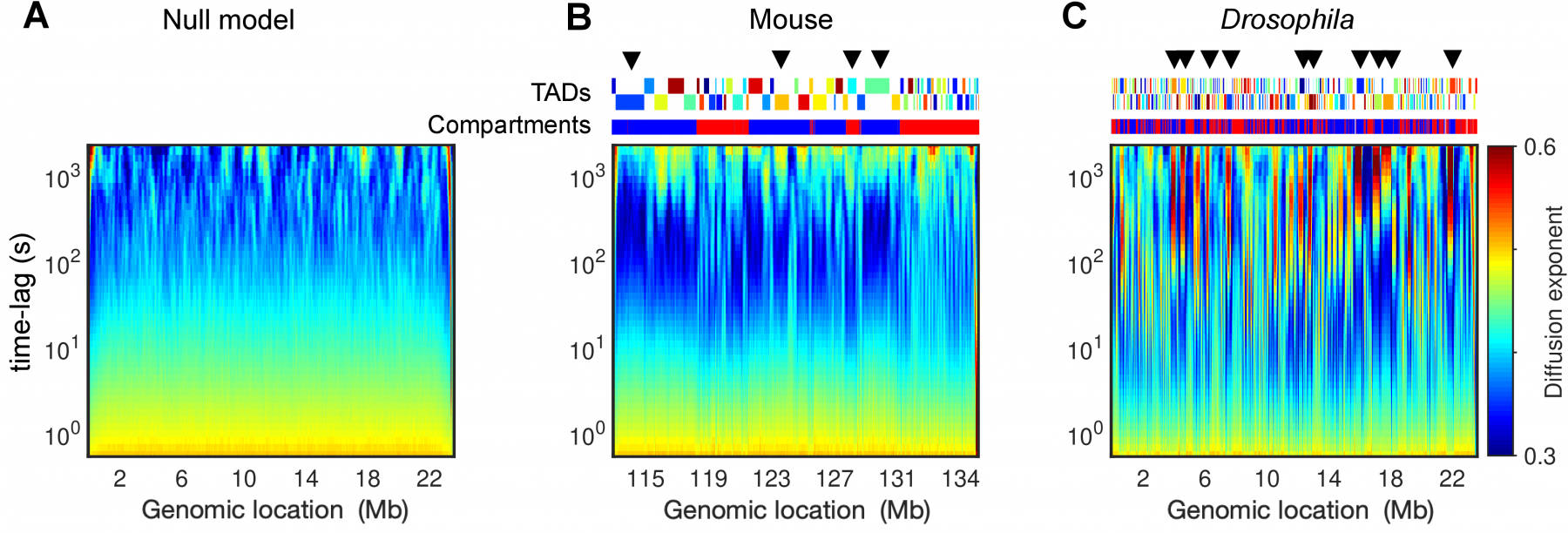
Chromosome organization and dynamics are involved in regulating many fundamental processes such as gene transcription and DNA repair. Experiments unveiled that chromatin motion is highly heterogeneous inside cell nuclei, ranging from a liquid-like, mobile state to a gel-like, rigid regime. Using polymer modeling, we investigate how these different physical states and dynamical heterogeneities may emerge from the same structural mechanisms. We found that the formation of topologically associating domains (TADs) is a key driver of chromatin motion heterogeneity. In particular, we showed that the local degree of compaction of the TAD regulates the transition from a weakly compact, fluid state of chromatin to a more compact, gel state exhibiting anomalous diffusion and coherent motion. Our work provides a comprehensive study of chromosome dynamics and a unified view of chromatin motion enabling interpretation of the wide variety of dynamical behaviors observed experimentally across different biological conditions, suggesting that the “liquid” or “solid” state of chromatin are in fact two sides of the same coin.
Download “Article preprint” Spatial_organization_of_chromosomes.pdf – Downloaded 219 times – 7 MB
Download a copy of the manuscript (preprint version)