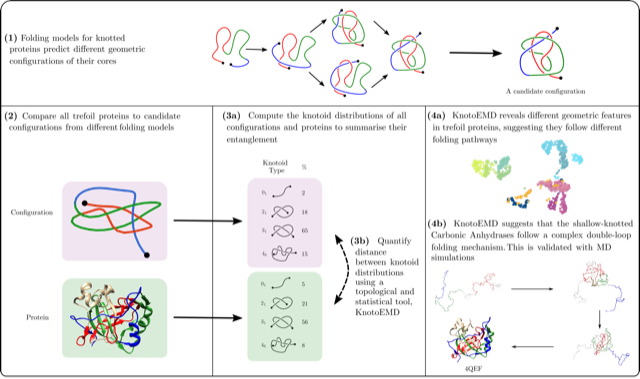
Understanding how knotted proteins fold is a challenging problem in biology. Researchers have proposed several models for their folding pathways, based on theory, simulations and experiments. The geometry of proteins with the same knot type can vary substantially and recent simulations reveal different folding behaviour for deeply and shallow knotted proteins. We analyse proteins forming open-ended trefoil knots by introducing a topologically inspired statistical metric that measures their entanglement. By looking directly at the geometry and topology of their native states, we are able to probe different folding pathways for such proteins. In particular, the folding pathway of shallow knotted carbonic anhydrases involves the creation of a double-looped structure, contrary to what has been observed for other knotted trefoil proteins. We validate this with Molecular Dynamics simulations. By leveraging the geometry and local symmetries of knotted proteins’ native states, we provide the first numerical evidence of a double-loop folding mechanism in trefoil proteins.
Download “Article” A_Topological_Selection_Folding_Pathways.pdf – Downloaded 251 times – 8 MB
Download a copy of the manuscript