Status
Completed
Period
XX-XX XXX 2022
Applicant
Prof. Samuela Pasquali
Home Institution
School of Pharmacy and Biology, Université Paris Cité
Host Contact
Prof. Pietro Faccioli
Host Institution
Physics Department, Università di Trento
Aim of the mission
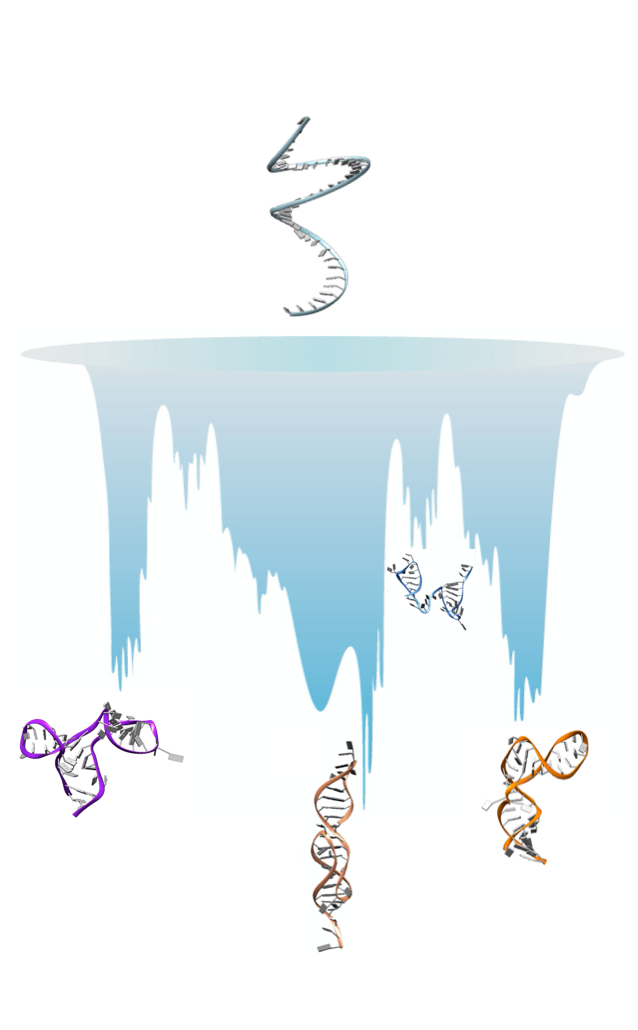
Non-coding RNA molecules have become the center of attention in biophysical and medical research because of their central role in the regulation of gene expression. With only a few percent of the genome of higher organisms coding for proteins, RNA constitutes an important target for a whole new family of drugs. Just like proteins, these molecules need to adopt specific three-dimensional shapes to perform their biological function. RNA sequences however have the peculiar feature of being able to fold on several distinct architectures, with alternative overall organizations. For an RNA molecule this organization is mainly guided by the hydrogen bond network of base pairing, which gives rise to the secondary structures. Secondary structures, in turns, can be classified in families of similarity, according to their topology. Indeed, using a description typical of graph theory, the organization in loop and stems can be mapped onto a graph and topological features can be assigned to it. Therefore, the study of RNA architectures is intrinsically related to the topology of its graph representation. The central interest of our group is to understand the structures adopted by RNA molecules, their stabilities, and their interconversion. To this purpose we develop simulation methods using force-field design and enhanced simulation techniques. Path-sampling simulations developed by Pr. Faccioli at the University of Trento are particularly promising for the study of RNA and the exploration of their alternative topologies. This new kind of simulation can follow the folding of biomolecules onto a target state, proposing insights on the folding mechanism. It can study large systems that can exhibit a variety of folds, including knots, as it was done in the past for proteins. A particularly interesting feature of such a method is the ability to identify transient states. This is very relevant for RNA systems, where the transient states can become dominant according to environmental conditions. Over the last few months, together with Pr. Faccioli, we have started investigating the application of this path-sampling method to RNA, starting with some systems for which we have extensive experience, having used them as benchmark for our force-field developments in the past. Among them are systems including pseudoknots, which are characterized by a rich folding pathway, as remarked also in studies using other methods to explore the energy landscape of these systems. This Short Term Scientific Mission will give the opportunity to Pr. Faccioli to finalize the first project on the proof of principle of the application of the path-sampling simulations to RNA. At this stage it is important for us to discuss in person the results of simulations, looking at trajectories and graphs on the same screen, which can hardly be done remotely. As a result of the STSM we plan to submit an article for publication, also including two other researchers (C. Micheletti at SISSA and Gianmarco Lazzeri at Frankfurt Institute for Advanced Studies). The mission will also give us the opportunity to define a future strategy for our collaboration. After this first work, we want to apply this method to the study of systems characterized by complex topologies that can hardly be tackled by other methods but that are at the heart of attention in the nucleic acids community. Two such systems are Guanine quadruplexes, formed by both DNA and RNA that are considered a
primary target for cancer drugs, and regulatory pseudoknots such as the frameshifting element of SARS-Cov-2.
Proposed contribution to the action
Because the study of RNA is a much younger field than that of DNA and protein, questions about the connections between their folding and their topology have not yet been addressed in an organized and comprehensive manner. This reflects in the fact that in Eutopia the two working groups dealing with biomolecules focus mainly on proteins and on DNA, and only minimal aspects of RNA systems are considered. Our work will allow to include RNA systems in Eutopia’s radar and will be the first seed for larger collaborations including other Eutopia’s member, which, with time, will
result in a network of researchers devoted to this topic. During this STSM we plan to discuss also with other Eutopia members that will be in
Trento at the same time to plan a workshop on RNA topology to be held in 2022.
Techniques
Path-sampling simulation of 3 RNA molecules have been performed by Pr. Faccioli uses the ratchet technique, consisting of applying an unfolding force bias based on a target structure during a molecular dynamic simulation starting from an unfolded system. With this technique the molecule evolves freely but when native contacts are
formed, they are preserved by the biasing force that forms a barrier such that the molecule does not unfold. Simulations have been performed on the computing cluster in Trento and are now in the process of being analyzed both by Pr. Faccioli, using tools developed specifically to monitor these kinds of simulations, and by Pr. Pasquali, using tools specific to the analysis of RNA trajectories to monitor formation of secondary structures and topology evolution. During the STSM we will not foresee to run new simulation, but to continue the analysis combining the findings of both Pr. Faccioli and Pr. Pasquali.
Summary of the results
During my stay in Trento I was able to successfully carry out the goals of the mission. I worked with Pr. Faccioli on the analysis of the path sampling simulations of RNA molecules that were previously produced, and we made significant advancements in finalizing a manuscript on this work. During the week, we were also in constant contact with Pr. Micheletti at SISSA via zoom, who is also involved in this project. The work we have done up to now is a proof of principle of the feasibility of this kind of investigations for RNAs. Together with Pr. Faccioli and Pr. Tubiana, we discussed how to continue this work in the future and identified a class of systems that will be particularly interesting to investigate first. Among them are molecules of therapeutic interest and that have intricate topologies such as G-quadruplexes and the frameshifting pseudoknot of Sars-Cov-2. Together also with the director of studies there, Pr. Lattanzi, we discussed ways to involve master students in these projects, with the possibility of a co-direction of the M2 thesis, between the University of Trento and the University of Paris. In particular we identified possible ways of giving the students the necessary financial support. Lastly, together with Pr. Tubiana and Pr. Potestio, we discussed the training actions for
the remaining time of Eutopia, and in particular the practical organization of the training school that will take place in the summer of 2022. We also discussed the future after Eutopia, to see how we can best capitalize of the COST action and have it evolve into a training network.
Dissemination
One published paper:
“Computer-aided comprehensive explorations of RNA structural polymorphism through complementary simulation methods”, K Röder, G Stinermann, P Faccioli, S Pasquali, QRB Discovery, 1-22 (2022)
One submitted paper, deposited on the archive:
“RNA folding landscapes from explicit solvent all-atom simulations”, G Lazzeri, C Micheletti, S Pasquali, P Faccioli, arXiv preprint arXiv:2205.12603