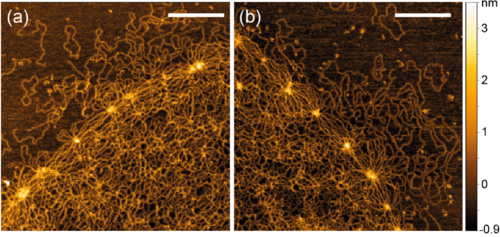
The Kinetoplast DNA (kDNA) is a two-dimensional Olympic-ring-like network of mutually linked 2.5 kb-long DNA minicircles found in certain parasites called Trypanosomes. Understanding the self-assembly and replication of this structure are not only major open questions in biology but can also inform the design of synthetic topological materials. Here we report the first high-resolution, single-molecule study of kDNA network topology using AFM and steered molecular dynamics simulations. We map out the DNA density within the network and the distribution of linking number and valence of the minicircles. We also characterise the DNA hubs that surround the network and show that they cause a buckling transition akin to that of a 2D elastic thermal sheet in the bulk. Intriguingly, we observe a broad distribution of density and valence of the minicircles, indicating heterogeneous network structure and individualism of different kDNA structures. Our findings explain outstanding questions in the field and offer single-molecule insights into the properties of a unique topological material.
Download “Article preprint” SingleMolecule_Kinetoplast.pdf – Downloaded 236 times – 8 MB
Download a copy of the manuscript (preprint version)