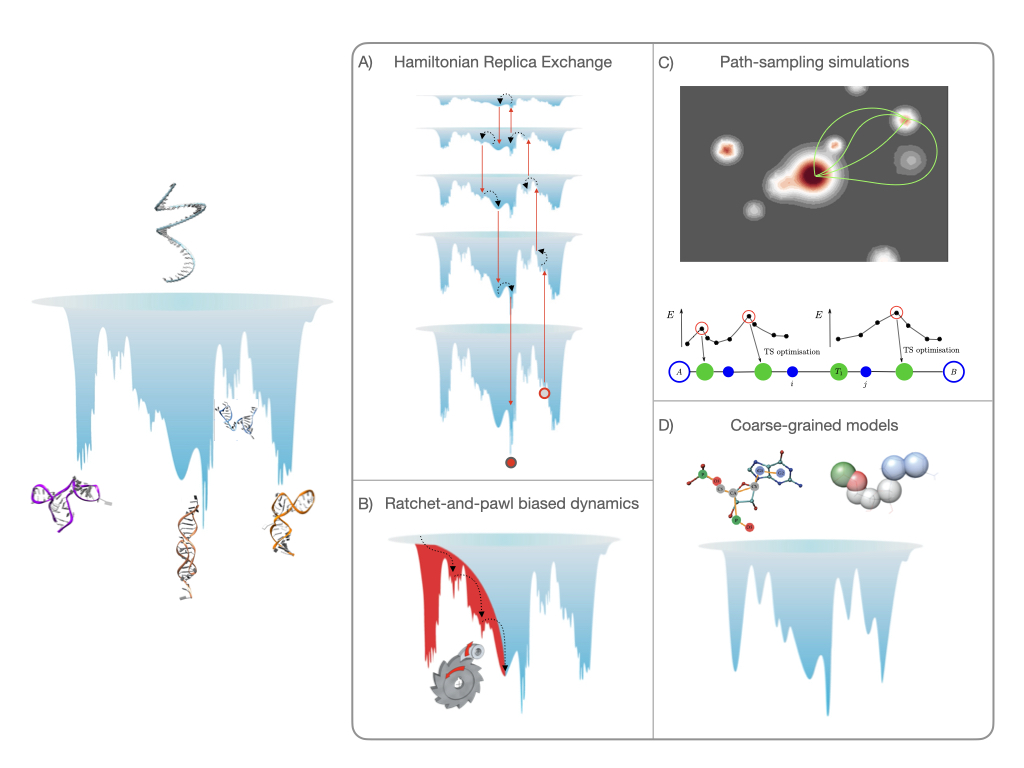
While RNA folding was originally seen as a simple problem to solve, it has been shown that the promiscuous interactions of the nucleobases result in structural polymorphism, with several competing structures generally observed for non-coding RNA. This inherent complexity limits our understanding of these molecules from experiments alone, and computational methods are commonly used to study RNA. Here, we discuss three advanced sampling schemes, namely Hamiltonian-replica exchange molecular dynamics, ratchet-and-pawl molecular dynamics and discrete path sampling, as well as the HiRE-RNA coarse-graining scheme, and highlight how these approaches are complementary with reference to recent case studies. While all computational methods have their shortcomings, the plurality of simulation methods
Download “Article” Computer_aided_comprehensive_explorations_RNA.pdf – Downloaded 186 times – 959 KB
Download a copy of the manuscript